Research
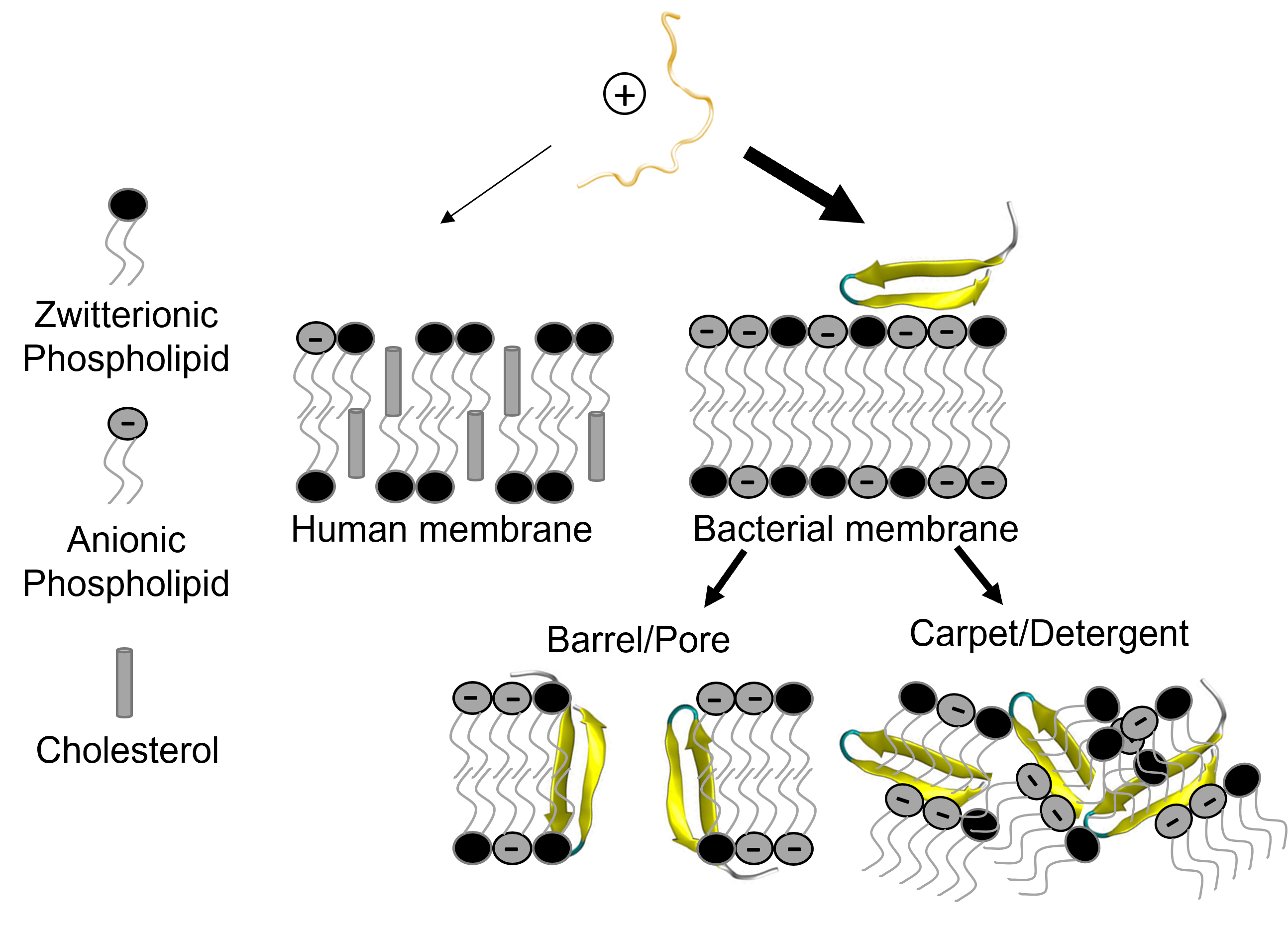
Manipulating membrane disruptive peptide cell specificity
Membrane disruptive peptides (MDPs) are a highly diverse peptide class found naturally throughout life in venoms, toxins, and host immune systems and carry a range of antimicrobial and anticancer activities. They also have a remarkable capacity to bind and disrupt membranes from diverse cell types with varying degrees of specificity. Their ability to bind and/or compromise cell membranes selectively makes them exciting candidates for therapeutic development as intracellular delivery vehicles, drugs directly targeting and killing pathogens/cancers, or for localized delivery of MDPs as live biotherapeutics. Unfortunately, a fundamental lack of information regarding what drives MDP cell membrane specificity continues to hinder their therapeutic use, often due to toxicity concerns.
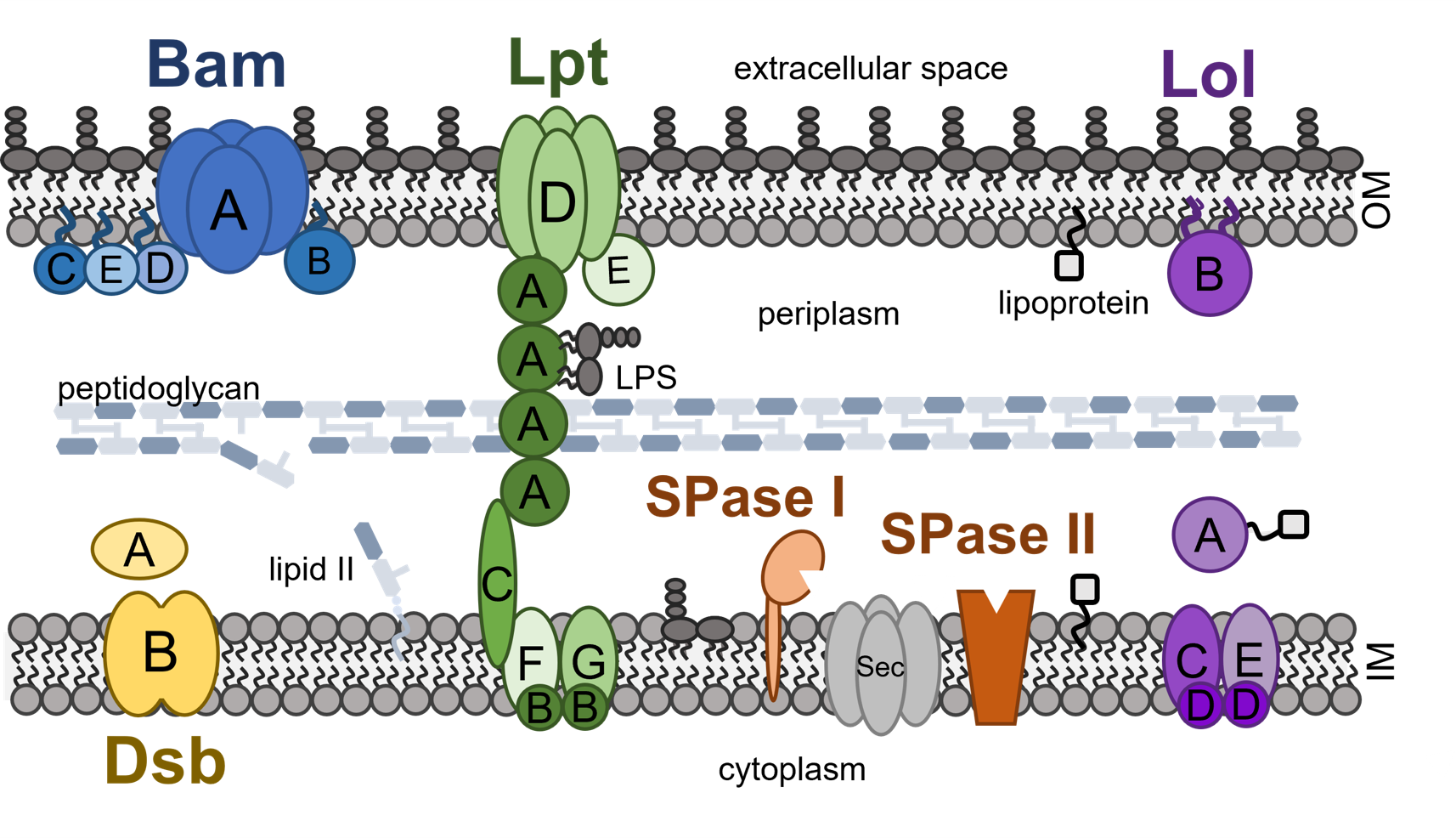
Finding peptide antibiotics targeting the bacterial cell envelope
We use a technique call surface localized antimicrobial display (SLAY) to screen hundreds of thousands of candidate peptides for inhibition of essential processes on the bacterial cell surface and within the bacterial cell envelope. These essential processes include membrane integrity, the beta barrel assembly complex (Bam), lipopolysaccharide transport (Lpt), lipoprotein transport (Lol), and periplasmic processes involved in disulfide bond formation (Dsb) and protein secretion (Sec/SPases). Once we identify lead candidates in our screens, we attempt to confirm their mechanism of action and determine their structure, antibacterial potency, and human toxicity.
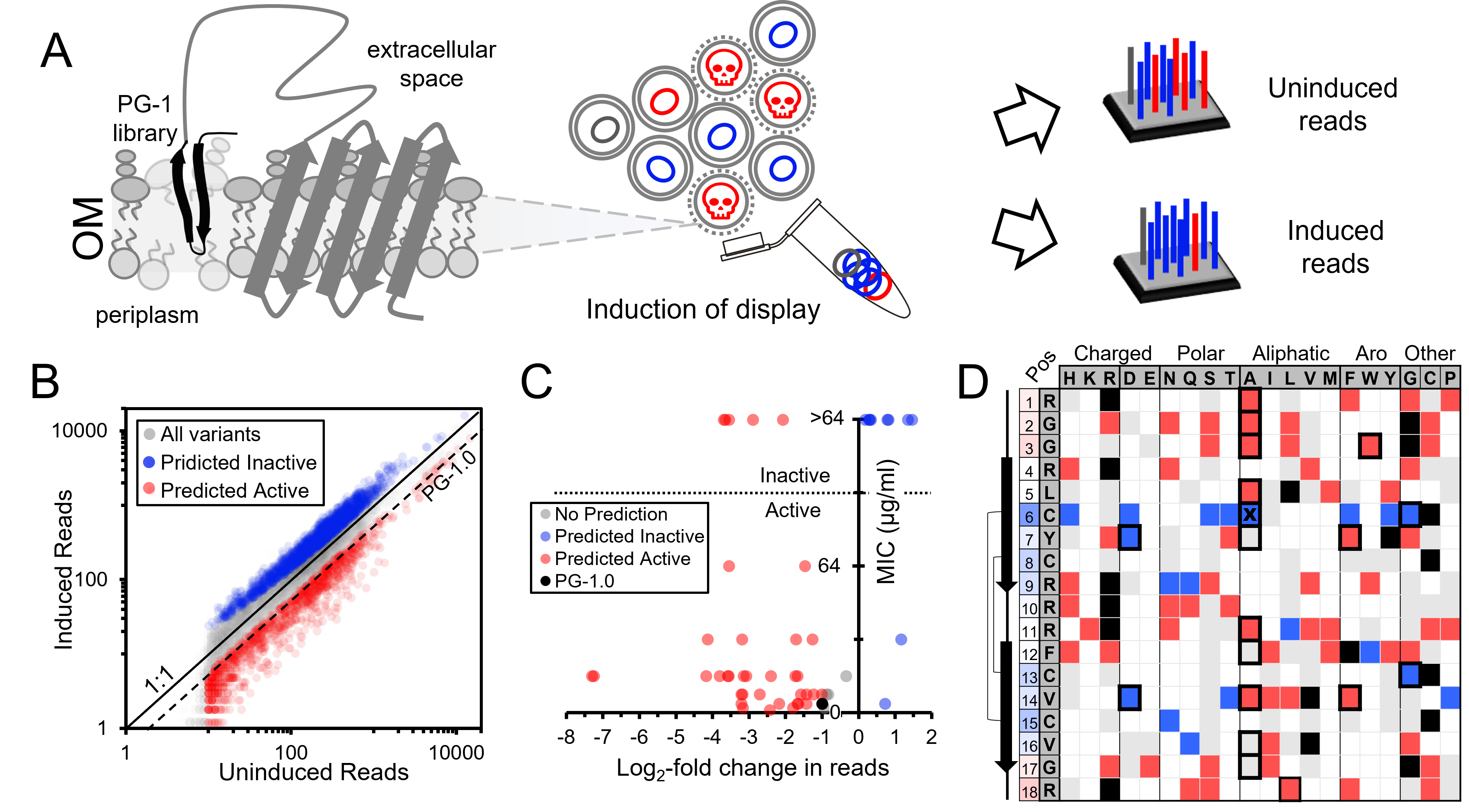
Developing SLAY screens for identification of antifungal and anticancer peptides
We are interested in developing surface localized antimicrobial display (SLAY) with fungal and tissue culture cells to help us identify antifungal and anticancer peptides. We are actively looking for students or staff with a background or interest in mycology and/or tissue culturing to help get these projects off the ground. If you are interested please contact us!
Recent Publications
For a full list of publications please visit my Google Scholar page
